Lec. 6. Biol C2005/F2401 2000 L. Chasin September 21, 2000
(C) Copyright 2000
Lawrence Chasin and Deborah Mowshowitz Department of Biological
Sciences Columbia University New York, NY
Last recorded update: Tuesday, September 26, 2000 09:15 PM
Exam #1 is Thursday evening, Sept. 28, 1999, at 7:00 or 5:40, depending on your class and/or last name. Click here for your room/time assignment.
There will be a review session (with Dr. Chasin) to answer your questions on Thursday 9:10 - 10:25 AM in 309 Havemeyer.
-----------------------------------------------------------------------------
Protein purification methods:
ultracentrifugation
native gel electrophoresis
SDS-gel electrophoresis
molecular sieve chromatography
Membrane proteins
Proteins bind molecules with great specificity
Protein domains
Catalysis
Activation energy
Substrates & products
Chemical kinetics
-------------------------------------------------------------------------------
(Protein purification
methods)
PROTEIN PURIFICATION/SEPARATION METHODS
(ultracentrifugation)
Here is one sometimes useful method: ULTRACENTRIFUGATION
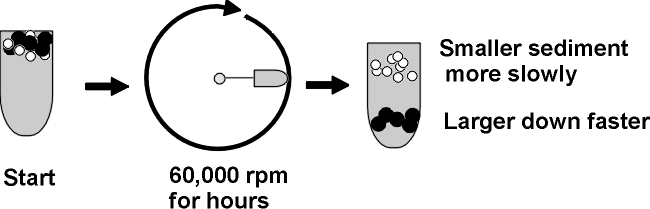
ULTRACENTRIFUGATION
Ultra means = >20,000 rpm; 60,000 rpm is common, compare a Ferrari revving at 6000 rpm, red-lining; this is ten times faster; you need a vacuum chamber so no heat from air friction will be produced.
Diagram of tube, spin, distribution of molecules ... ...

A mixture of molecules will be subject to two
main forces in the ultracentifuge as it starts to spin (ignoring buoyant force):
Causing sedimentation is the centrifugal
force = m(omega)2r = (which is proportional to the
mass or MW of a protein).
m = mass, omega = angular velocity, and r = distance from the
center of rotation.
Opposing sedimentation = friction = foV.
fo = frictional coefficient, a
constant for any particular protein, it is minimum for a sphere, higher for less
compact shapes like cigars or pancakes.
V = velocity of the molecule as it moves away from the center
of rotation .
Soon after accelerating, V increases to a point where no further acceleration takes place, as the forces on the molecule are balanced. It continues to sediment, but at a constant velocity.
Now at this point, at this velocity: Centrifugal Force = Frictional force (there's no net force, no acceleration, but constant velocity)
So at this point (soon achieved): M(omega)2r = foV
And: V = m(omega)2r/fo,
where f = a frictional coefficient dependent on
shape
(to visualize the effect of shape on friction, compare the velocity of a falling
feather vs. a tiny pebble of equal weight, dropped in the fluid of air).
Higher f = more friction.
If assume a spherical shape, then we can estimate a MW (Assume fo, and then measure V and r, so we can solve for m, or the MW)
On the other hand, if we know the MW, we can get information about shape (via fo).
Sedimentation velocity is often measured in Svedbergs, which takes the centrifugation conditions into account s = V/(omega)2r, and so m = sfo.
So ultracentrifugation separates proteins on the basis of MW and shape. It is a gentle procedure (non-denaturing, can be carried out at nice low temperature (say 4 deg C, which tends to stabilize proteins) and in the presence of a buffer at pH 7 and physiological levels of salts).You can recover your protein by punching a hole in the bottom of the centrifuge tube, and collecting the solution in a series of tubes as it drips out the bottom. Each tube can then be examined, or assayed, for the presence of the protein to be purified. For this purpose you need to be able to detect the protein in the midst of the other proteins. For example, if you were purifying Anfinsen's ribonuclease, you could measure the ability of the tube contents to catalyze the breakdown of RNA to its monomers.
How about separation on the basis of the net charge of a protein. We separated amino acids on the basis of charge in paper electrophoresis. For proteins, the solid supporting material is a gel, not paper:
GEL ELECTROPHORESIS:
There are two types -
( native gel electrophoresis)
First: native
gel electrophoresis
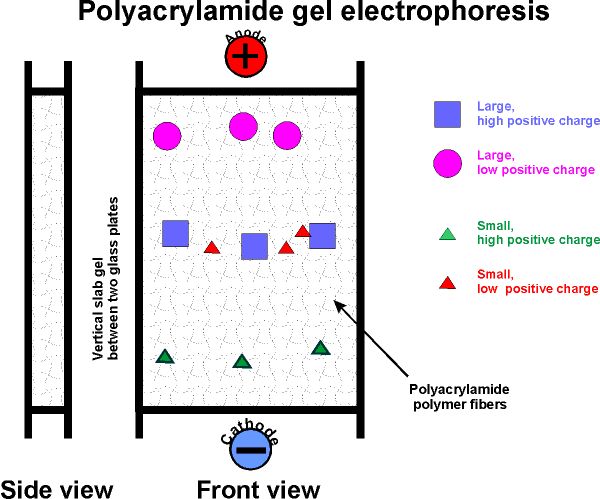
acrylamide, a reactive chemical (a monomer in this chemistry) in aqueous solution can be made to polymerize into polyacrylamide (thus polyacrylamide gel electrophoresis, or P.A.G.E.). The result is a network of polymer fibers, which form a gel, with about the consistency of Jello.
Usually carried out in an apparatus in which this gel stands vertically supported in a sandwich between two glass plates. The top of the gel is connected by a salt solution to an electrode, as is the bottom edge of the gel. A protein mixture is applied to the top of the gel slab.
The electrophoresis is started by applying a voltage (~200 v) across the gel.
The rate of migration of a protein in the gel
depends on two properties:
The first is its net charge.
Molecules with the most charge (of a sign opposite to that of the far electrode)
will migrate to the far electrode fastest.
Second, its "size" (which is
proportional to MW if spherical). The gel consists
of a network of fibers. Depending on the concentration of polyacrylamide, the
network canbe dense or tight, so that the proteins have trouble migrating, as
they must negotiate their way through the tangled fibers. Molecules that are
smallest (i.e., lowest MW) will worm their way through the gel fibers fastest.
So the smallest and most highly charged protein wins the race.
After the electrophoresis has been stopped, molecules will be distributed along
the gel length according to these two characteristics (MW and net charge).
For instance, a highly charged protein molecule, although pulled with a
greater electromotive force, will not have gotten very far if it is relatively
large.
[Note that molecules with a charge opposite to the near electrode, will migrate
up and off the gel, into the buffer reservoir and be lost. Trial and error
will dictate how you setup the electrophoresis if you do not know the charge on
the protein you are trying to isolate.]
Sometimes the gel is made purposely loose, so that the effective pore size is very large and then charge alone determines mobility. Starch gels rather than polyacrylamide have been used to create large pore gels.
(SDS gel electrophoresis)
A second, a
more widely used variation of gel electrophoresis is SDS
PAGE.
Here sodium dodecyl sulfate, or SDS (or SLS) is included in the gel: CH3-(CH2)11- SO4=
[sulfate is similar in structure to phosphate, and is a strong acid]. Like a phospholipid, SDS has a highly polar end and a highly hydrophobic body.
Might you expect SDS to denature a protein? Yes. It's a detergent and a powerful denaturant. It binds all over the protein, coating every protein with a nearly uniform negative charge. SDS is put into in the gel when you form it and into the electrophoresis buffer. Now when SDS-PAGE is run, where should the anode be placed? Does it matter? Yes, the protein is coated with negative charge now, so the anode is always the far electrode, put at the bottom.
Under these denaturing conditions, the polypeptides exist as a random coils, which then migrate solely on the basis of their size, which is the equivalent of a sphere for all polypeptides. Larger molecules have more difficulty finding their way through the polyacrylamide fibers. So the lowest MW wins always wins in SDS-PAGE.
If you run standards of known MW, you can determine the MW of your protein by comparison, and this is a very common way to assign a MW to a polypeptide. However, it is not always completely accurate, as some proteins probably do bind a bit more SDS than others. And one must remember to reduce the disulfides with mercaptoethanol first (usually), so as to have a truly random coil for comparisons with other proteins.
If you don't yet know what a protein does, you can just call it by its molecular weight, from SDS gels: e.g., p53, a famous protein whose absence is associated with cancer was named this way, and the name has stuck even though quite a lot is known about its function now (p in p53 stands for protein, so you have names like p27, p100 etc.).
(Gel filtration)
If you have a protein with quaternary
structure, SDS-PAGE will give you the MW of the polypeptide subunits, since the
SDS will denature the protein and so dissociate it into its subunits If
you want to know the MW of a protein in its native quaternary structure, you
need a method seprates proteins under mild conditions that maintain its native
structure.
For this we could use molecular sieve chromatography, also called or Sephadex chromatography , or gel filtration (these are all ~synonymous).
You start with plastic-like beads in a glass column with a support screen on the bottom.
You add your protein mixture to the top of the column material, then begin elution by adding a large volume of a buffer. The beads have been manufactured to be riddled with channels of a specified fairly uniform size. If a protein is smaller than the channel size, it enters a channel, explores it by diffusion, and eventually makes its way back out, having wasted its time in the race to the bottom of the column. Larger proteins can't fit in to the channels, so they don't waste their time, and they win the race. Intermediate sized proteins have a bit of trouble getting into a channel, so they waste some time but less than the smaller proteins. So larger molecules come out (elute) first, and the smallest come out last. Here again, you would collect the eluted proteins in a series of tubes, and then assay each tube for the presence of the protein being purified. If you calibrate the column by noting the behavior of spherical proteins of known size, you can determine the MW of your protein by comparison, if it is also spherical. If is is not spherical it will appear to have a higher molecular weight than its true MW (imagine a pancake being excluded from a channel while a sphere of the same MW gets in).
Other methods include ion exchange chromatography, which also takes advantage if the net charge on a protein, and affinity chromatography, which takes advantage of the surface properties of a protein (which we'll discuss next). One can purify a particular protein away from all other proteins in 4-5 such steps. For more on these protein separation techniques, see the protein separation handout.
Here are some images of laboratory ultracentrifuge , slab gel electrophoresis, gel filtration apparatuses.
One has to able to follow the protein of interest, to detect its presence in the presence of all others. Often use its functional properties for this, which brings us back to structure and function of proteins.
(Membrane proteins)
As an introduction into an example of the
function of proteins, let's consider first a special class of proteins that do
not follow some of the rules we have have seen that govern protein structure in
general. These are the MEMBRANE PROTEINS, the
proteins that reside, not in the aqueous environment of the cytoplasm or the
nucleus, but rather in the hydrophobic environment of the membranes of the cell,
including the cell membrane.
In these proteins, the hydrophobic side chains are on the outside, as there are
no hydrophobic forces present i to force then to coalesce. These proteins can
usually diffuse laterally in the lipid bilayer of the membrane [Purves 5.4],
they can aggregate with each other with specificity, and they can become anchored
via attachment to structural cytoplasmic fibers. They can be nearly
completely enveloped by the lipid bilayer, or they could be partially immersed,
with a more conventional half (i.e., hydrophobics on the inside, hydrophilics on
the outside) sticking out into the cytoplasm or on the outside of the
cell.
See [Purves 5.1], [Purves 5.5].
One class of membrane proteins act as channels through the membrane. The
channel proteins are formed into cylinders, with a hydrophobic exterior, but
with hydrophilic groups lining the hole through the interior of the cylinder.
Small molecules can pass through the cylinder, or channel, but large molecules
cannot fit through (note: macromolecules CANNOT diffuse onto cells).
So we have a cellular function for a protein, and a function that shows some selectivity on the basis of size there. How about
other criteria of selectivity, like charge?
Some channels can distinguish charge: + repels +, and attracts -, so a channel that is lined with positive charge could bind a negatively charged ion and, if there's a higher concentration outside than inside, eventually pass these ions along from the outside of the cell to the inside. A positively charged ion on the other hand would be repelled by the channel and so would not get into the cell by this route.
You can a imagine the same sort of selectivity based on hydrogen bonding, for example.
So a protein can detect charge and surface electrical properties, but how about shape?(Proteins bind molecules with great specificity)
Consider a pocket on the surface of a folded protein.
As I've drawn this surface pocket, the free amino acid gly can fit in and bind
using electrical attraction or ionic bonds. However, the closely related amino
acid alanine, with a methyl group as a side chain, cannot fit into this hypothetical protein A drawn here
(yellow). Ionic bonds and van der Waals bonds (VDW) are responsible for the binding in this example so
far (using different colors for protein and aa)
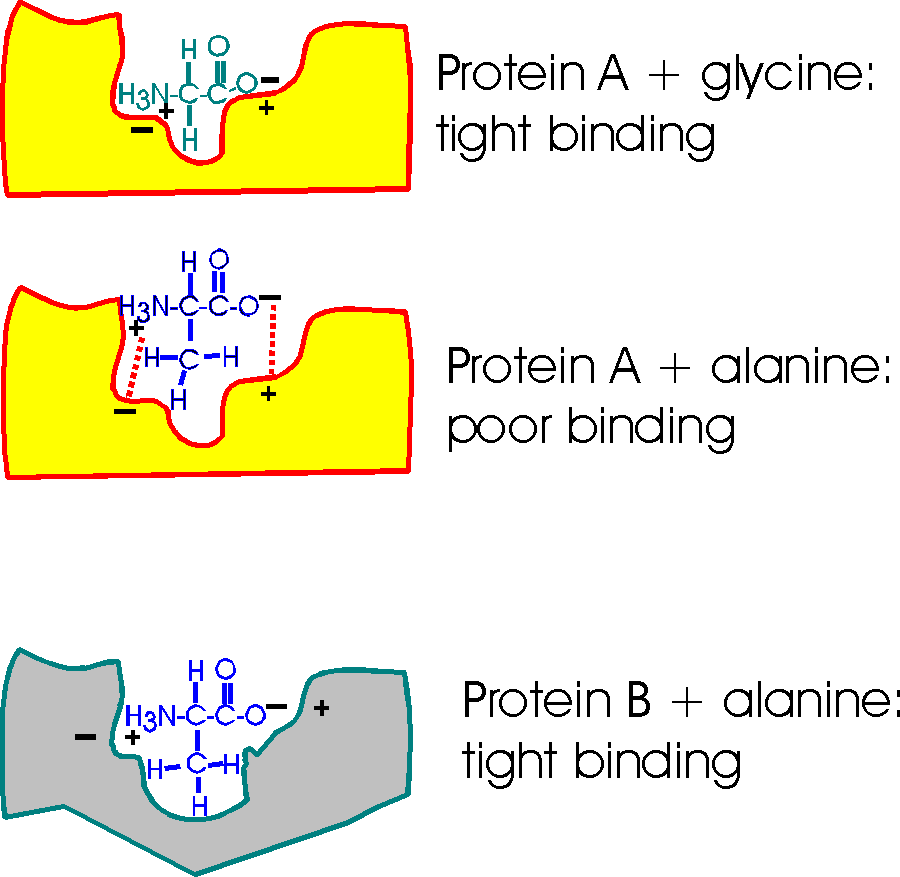
But a similar pocket on the surface of another protein (gray protein B) could be built to accommodate the methyl (or -CH3 group) of alanine and supply some more V.D.W. bonds there in the process.
So Protein A binds glycine, but not alanine.
And Protein B binds alanine (but gly not so tightly).
So you can get specific binding ... and this binding is critically dependent upon the structure of the protein, the shape of the binding site. This binding is also a critical part of the function of the protein, and we will soon see.
Specific binding at a protein surface is not restricted to interactions between a macromolecule and a small molecule. There is also specificity in the interactions between two macromolecules, as exemplified many times by quaternary structure: the complementary surfaces of the two correct subunits fit together with great specificity; just the right subunits polypeptide specifically associate to form a multimeric protein. For example, the subunits of immunoglobulin (Ig) never associate with the subunits of hemoglobin (Hb).
(Domains)
DOMAINS: Now that we have seen that proteins can bind
other molecules with great specificity, I should mention one last additional aspect of
protein structure that I have put off until now: protein domains. The overall shape of most proteins is
roughly globular, but if one looks more closely, one can see that most proteins can be
divided into sub-regions that are folded more or less independently of the rest. These
folded up globules are called domains. An interesting feature of many domains is that
homologous domains can often be found in many different proteins. Many of the
individual amino acids in the primary structure are different, but many others are the same, and the overall shape
of the domains in different proteins can be very similar. A recognizable domain in a protein can often be associated
with a particular function, often the ability to bind a particular small molecule.
In the simple example we used above, a glycine-binding domain might be found in several
different proteins, each of which needs to bind glycine to carry out its function. Thus we
might also have a glucose binding domain, a phosphate-binding domain or an RNA-binding
domain in several proteins whose function requires them to bind these molecules.
ENZYMES
The ability to bind a specific small molecule is exploited by proteins when they carry out one their main functions: to act as catalysts that bring about chemical transformations of the small molecules they bind. These protein catalysts are called enzymes. Enzymes represent perhaps the single largest category of proteins, with respect to function. Since they are responsible for virtually all the chemical conversions going on in the cell, it is difficult to overestimate the central role they play in life.
(Catalysis)
Enzymes function as catalysts.
So let's define a catalyst.
Consider the purely chemical reaction between hydrogen gas and iodine gas:
H2 + I2 --> 2 HI + energy
This reaction goes spontaneously to the right because H2 and I2 are higher energy compounds than HI. That is, H2 and I2 are less stable than the combination of these 4 atoms in the form 2HI) :
[In the energy diagram below, the ordinate (y-axis) is free energy of the components, change in free energy (delta G) is only thing that can be measured, and free energy here is the energy needed to pull apart the atoms (highest bond strengths will be lowest on the ordinate, as it will mean more energy has to be put in to raise the atoms to their free, separated, state)].

SO if you could invest the energy to separate the atoms, and then let them fall back to HI's, you would get more energy out (3 kcal/mole difference). This is a characteristic of a spontaneous chemical reaction: spontaneous means the reaction can proceed in the direction indicated (left to right) with the release of energy. In contrast, reactions that do not release energy, but require energy input, are not spontaneous.
ENERGY RELEASING reactions are called EXERGONIC. ENERGY-REQUIRING reaction are ENDERGONIC. {Q&A} See [Purves 6.5]
Despite the fact that this is an exergonic reaction, it does not proceed very readily.
This failure to react
is the case for many such energy releasing reactions: e.g., burning paper (cellulose + oxygen
reaction) can release much energy, but left to itself in air paper only slowly
browns.
We can understand this failure to react if you consider that you'd need to get the atoms apart before you can rearrange them, and it takes a lot of energy to break those covalent bonds. Actually, you do NOT need to take the atoms completely apart:
(Activation energy)
To get this transformation to proceed, you just need to get to
a transition state. If the two molecules (H2 and
I2) collide at a sufficiently high velocity, then all
four of the atoms involved in the collision can 4 can temporarily form bonds to
each other, and this complex then has a chance to resolve itself into 2 HI (or back
in to H2 + I2):

So or the reaction to proceed, you only need to produce a transition state, and the energy needed to get to a transition state is called the ACTIVATION ENERGY.
See [Purves 6.11a], [Purves 6.11b], and [Purves 6.12].
CATALYSTS ACT BY REDUCING THE ACTIVATION ENERGY. See [Purves 6.14]. Without a catalyst, you need a forceful collision to get to the TS. Very few molecules can muster it. But, for our reaction here, if we add a third substance, if we add some powdered platinum, the reaction proceeds almost instantly. The platinum can bind both reactants, so that many of the hydrogen and iodine gas molecules find themselves as neighbors on the surface of the platinum. More like bedfellows, as they can be closely packed. So closely, that they can form a transition state right there on the surface of the platinum particle:

The platinum makes it easier to get to a transition state, no forceful collision is required, the two participants (reactants) just bind close together on the common binding surface. And binding to the Pt also weakens the H-H bond and the I-I bond, making it easier to now form the H-I bond. The CATALYST IS NOT ALTERED, it just speeds up the reaction. The catalyst does not change the situation with respect to the spontaneity of the reaction (energy releasing character, or DIRECTIONALITY, refer back to energy diagram), it just speeds things up.
[See animation (2 MB file though... probably not worth the wait unless you are connected to the CU network)].
Chemical catalysts such as Pt can speed things up 10,000 fold, so they are important in the chemical industry.
(C) Copyright 2000 Lawrence Chasin and Deborah Mowshowitz Department of Biological Sciences Columbia University New York, NY